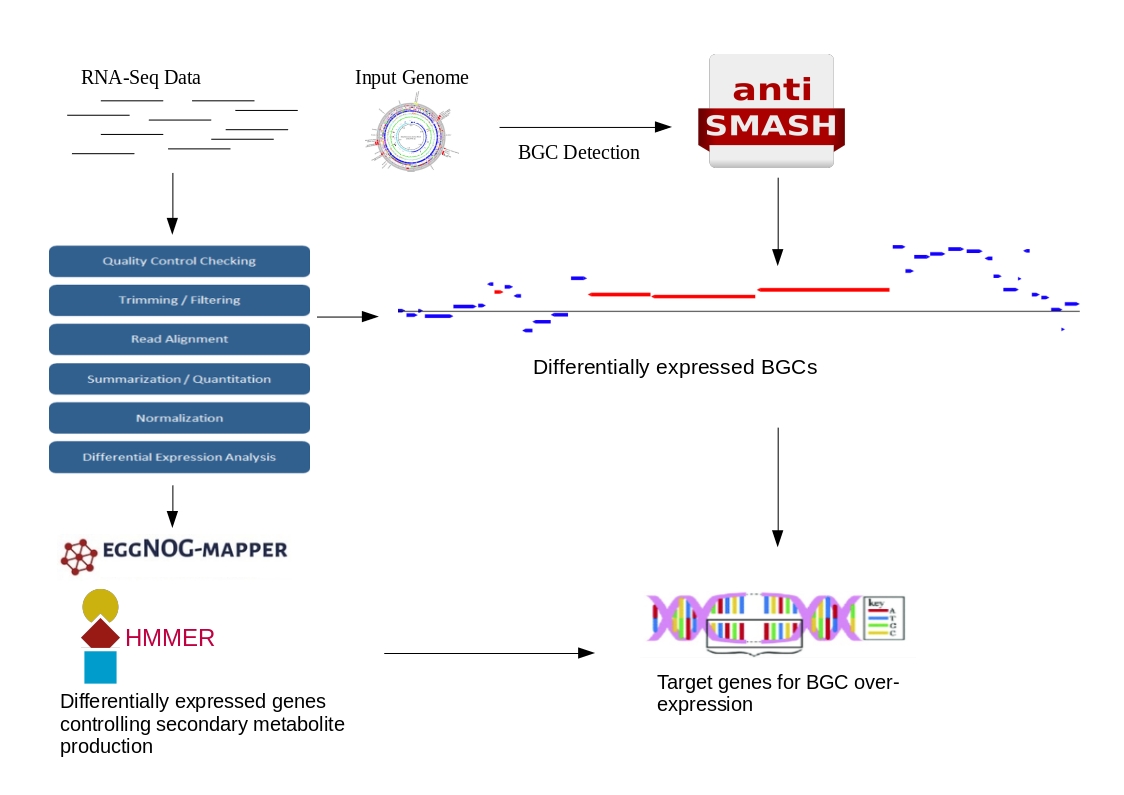
Sema-Trap is an analysis web-server/standalone tool for efficient prioritization of experiment design and target genes that can be used to over-express or activate biosynthetic gene clusters.
Using SeMa-Trap, researchers can:
- Select an experimental design based on BGC expression. To that aim, users can supply SRA run accession numbers from single condition experiment (e.g. control or treated) and check whether their BGC of interest is expressed based on housekeeping genes' expression. Afterwards, a full scale comparative transcriptomic experiment can be designed.
- Prioritize target genes for BGC over expression. For this purpose, a comparative experiment analysis can be made. Researchers can supply SeMa-Trap with either SRA run accession numbers or already analyzed&sorted "BAM" files. Afterwards, SeMa-Trap will return an overview page of the expressions of BGCs with respect to the provided experimental data. If there's an interesting BGC in terms of expression and fold change, an interactive result page for each BGC can be explored with specific target genes whose expression changes are connected to BGCs of interest.
SeMa-Trap is freely available to all users and there is no login requirement. The code is open source, licensed under the GPLv3 and stored through bitbucket. All third-party applications included are unrestrictive and free to use.